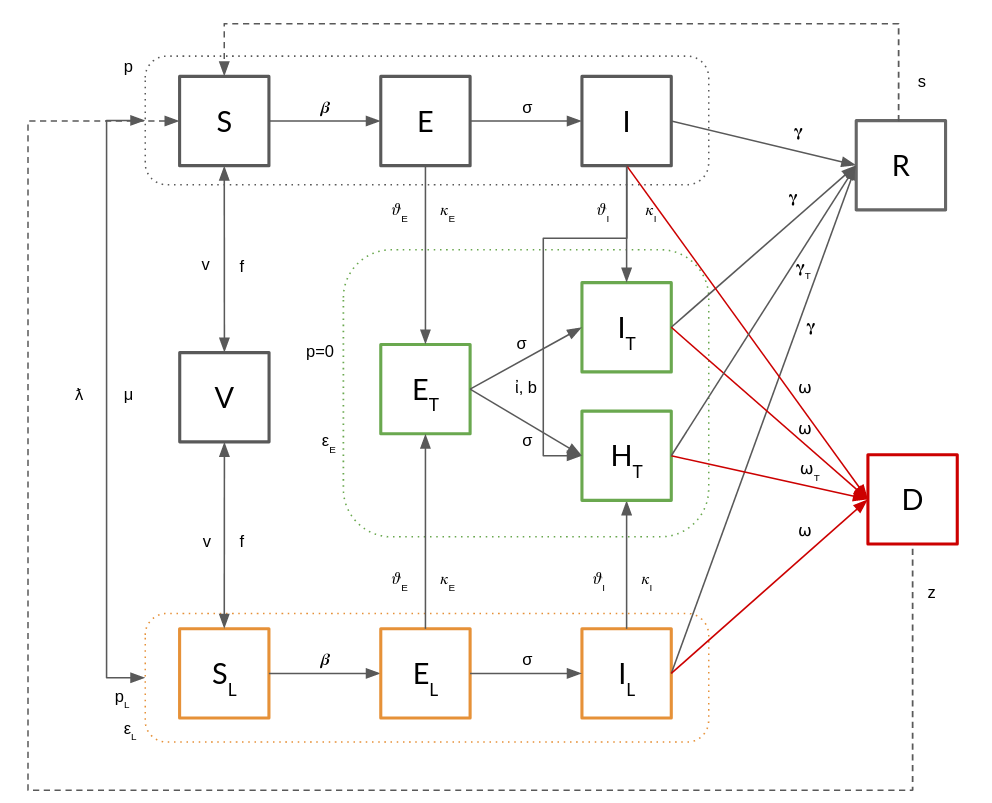
UTLDR¶
The UTLDR model [1] describe a complex framework allowing to extend SEIR/SEIS to incorporate medical and non medical interventions. The acronym summarizes the five macro statuses an agent can be involved into:
- U ndetected
- T tested
- L ocked
- D ead
- R ecovered
The U macro status follows the same rules of a classic SEIR(S) model and model the general epidemic evolution when no intervention is established.
The T macro status implements Testing (e.g., identification of exposed/infected agents) and model different classes of response (e.g., quarantine, hospitalization, ICU ospitalization).
The L macro status implements Lockdowns (that can be fine tuned on node attributes) and, as such, social contacts reduction.
Finally, the R and D statuses model the final outcome of an infection (either death or recovery) and are sensible to the various paths for reaching them.
Moreover, UTLDR allows also to include (effective/uneffective) vaccination effects.
Statuses¶
During the simulation a node can experience the following statuses:
| Name | Code |
|---|---|
| Susceptible | 0 |
| Infected | 1 |
| Exposed | 2 |
| Recovered | 3 |
| Identified Exposed | 4 |
| Hospitalized Mild conditions | 5 |
| Hospitalized in ICU | 6 |
| Hospitalized in Severe conditions (not ICU) | 7 |
| Lockdown Susceptible | 8 |
| Lockdown Exposed | 9 |
| Lockdown Infected | 10 |
| Dead | 11 |
| Vaccinated | 12 |
Parameters¶
| Name | Type | Value Type | Default | Mandatory | Description |
|---|---|---|---|---|---|
| sigma | Model | float in [0, 1] | True | Incubation rate | |
| beta | Model | float in [0, 1] | True | Infection rate | |
| gamma | Model | float in [0, 1] | True | Recovery rate (Mild, Asymtomatic, Paucisymtomatic) | |
| gamma_t | Model | float in [0, 1] | 0.6 | False | Recovery rate (Severe in ICU) |
| gamma_f | Model | float in [0, 1] | 0.95 | False | Recovery rate (Severe not ICU) |
| omega | Model | float in [0, 1] | 0 | False | Death probability (Mild, Asymtomatic, Paucisymtomatic) |
| omega_t | Model | float in [0, 1] | 0 | False | Death probability (Severe in ICU) |
| omega_f | Model | float in [0, 1] | 0 | False | Death probability (Severe not ICU) |
| phi_e | Model | float in [0, 1] | 0 | False | Testing probability (if Exposed) |
| phi_i | Model | float in [0, 1] | 0 | False | Testing probability (if Infected) |
| kappa_e | Model | float in [0, 1] | 0.7 | False | 1 - False Negative probability (if Exposed) |
| kappa_i | Model | float in [0, 1] | 0.9 | False | 1 - False Negative probability (if Infected) |
| epsilon_e | Model | float in [0, 1] | 1 | False | Social restriction, percentage of pruned edges (Quarantine) |
| epsilon_l | Model | float in [0, 1] | 1 | False | Social restriction, percentage of pruned edges (Lockdown) |
| lambda | Model | float in [0, 1] | 1 | False | Lockdown effectiveness |
| mu | Model | float in [0, 1] | 1 | False | Lockdown duration (1/expected iterations) |
| p | Model | float in [0, 1] | 0 | False | Probability of long range (random) interactions |
| p_l | Model | float in [0, 1] | 0 | False | Probability of long range (random) interactions (Lockdown) |
| lsize | Model | float in [0, 1] | 0.25 | False | Percentage of long range interactions w.r.t. short range ones |
| icu_b | Model | int in [0, inf] | N | False | ICU beds availability |
| iota | Model | float in [0, 1] | 1 | False | Severe case probability (requiring ICU) |
| z | Model | float in [0, 1] | 0 | False | Probability of infection from corpses |
| s | Model | float in [0, 1] | 0 | False | Probability of no immunization after recovery |
| v | Model | float in [0, 1] | 0 | False | Vaccination probability (once per agent) |
| f | Model | float in [0, 1] | 0 | False | Probability of vaccination nullification (1/temporal coverage) |
| activity | Node | float in [0, 1] | 1 | False | Node’s interactions per iteration (percentage of neighbors) |
| segment | Node | str | None | False | Node’s class (e.g., age, gender) |
The initial infection status can be defined via:
- fraction_infected: Model Parameter, float in [0, 1]
- Infected: Status Parameter, set of nodes
The two options are mutually exclusive and the latter takes precedence over the former.
Methods¶
The following class methods are made available to configure, describe and execute the simulation:
Configure¶
-
class
ndlib.models.epidemics.UTLDRModel.UTLDRModel(graph, seed=None)¶
-
UTLDRModel.__init__(graph)¶ Model Constructor
Parameters: graph – A networkx graph object
-
UTLDRModel.set_initial_status(self, configuration)¶ Set the initial model configuration
Parameters: configuration – a `ndlib.models.ModelConfig.Configuration`object
-
UTLDRModel.reset(self)¶ Reset the simulation setting the actual status to the initial configuration.
Describe¶
-
UTLDRModel.get_info(self)¶ Describes the current model parameters (nodes, edges, status)
Returns: a dictionary containing for each parameter class the values specified during model configuration
-
UTLDRModel.get_status_map(self)¶ Specify the statuses allowed by the model and their numeric code
Returns: a dictionary (status->code)
Execute Simulation¶
-
UTLDRModel.iteration(self)¶ Parameters: node_status – Returns:
-
UTLDRModel.iteration_bunch(self, bunch_size)¶ Execute a bunch of model iterations
Parameters: - bunch_size – the number of iterations to execute
- node_status – if the incremental node status has to be returned.
- progress_bar – whether to display a progress bar, default False
Returns: a list containing for each iteration a dictionary {“iteration”: iteration_id, “status”: dictionary_node_to_status}
Dynamically Update Policies¶
-
UTLDRModel.set_lockdown(self)¶ Impose the beginning of a lockdown
Parameters: households – (optional) dictionary specifying the households for each node <node_id -> list(nodes in household)> Returns:
-
UTLDRModel.unset_lockdown(self)¶ Remove the lockdown social limitations
Returns:
-
UTLDRModel.add_ICU_beds(self, n)¶ Add/Subtract beds in intensive care
Parameters: n – number of beds to add/remove Returns:
Example¶
In the code below is shown an example of instantiation and execution of an UTLDR simulation on a random graph.
import networkx as nx
import numpy as np
import ndlib.models.ModelConfig as mc
import ndlib.models.epidemics as epd
# Network topology
g = nx.erdos_renyi_graph(1000, 0.1)
model = epd.UTLDRModel(g)
config = mc.Configuration()
# Undetected
config.add_model_parameter("sigma", 0.05)
config.add_model_parameter("beta", {"M": 0.25, "F": 0})
config.add_model_parameter("gamma", 0.05)
config.add_model_parameter("omega", 0.01)
config.add_model_parameter("p", 0.04)
config.add_model_parameter("lsize", 0.2)
# Testing
config.add_model_parameter("phi_e", 0.03)
config.add_model_parameter("phi_i", 0.1)
config.add_model_parameter("kappa_e", 0.03)
config.add_model_parameter("kappa_i", 0.1)
config.add_model_parameter("gamma_t", 0.08)
config.add_model_parameter("gamma_f", 0.1)
config.add_model_parameter("omega_t", 0.01)
config.add_model_parameter("omega_f", 0.08)
config.add_model_parameter("epsilon_e", 1)
config.add_model_parameter("icu_b", 10)
config.add_model_parameter("iota", 0.20)
config.add_model_parameter("z", 0.2)
config.add_model_parameter("s", 0.05)
# Lockdown
config.add_model_parameter("lambda", 0.8)
config.add_model_parameter("epsilon_l", 5)
config.add_model_parameter("mu", 0.05)
config.add_model_parameter("p_l", 0.04)
# Vaccination
config.add_model_parameter("v", 0.15)
config.add_model_parameter("f", 0.02)
nodes = g.nodes
ngender = ['M', 'F']
work = ['school', 'PA', 'hospital', 'none']
for i in nodes:
config.add_node_configuration("activity", i, 1)
config.add_node_configuration("work", i, np.random.choice(work, 2))
config.add_node_configuration("segment", i, np.random.choice(ngender, 1)[0])
model.set_initial_status(config)
iterations = model.iteration_bunch(10)
households = {0: [1, 2, 3, 4], 5: [6, 7]}
model.set_lockdown(households, ['PA', 'school'])
iterations = model.iteration_bunch(10)
model.unset_lockdown(['PA'])
iterations = model.iteration_bunch(10)
model.set_lockdown(households)
iterations = model.iteration_bunch(10)
model.unset_lockdown(['school'])
iterations = model.iteration_bunch(10)
model.add_ICU_beds(5)
iterations = model.iteration_bunch(10)
| [1] |
|